|
sub-002 |
afni_proc.py single subject report
subj: sub-002
task: task_name
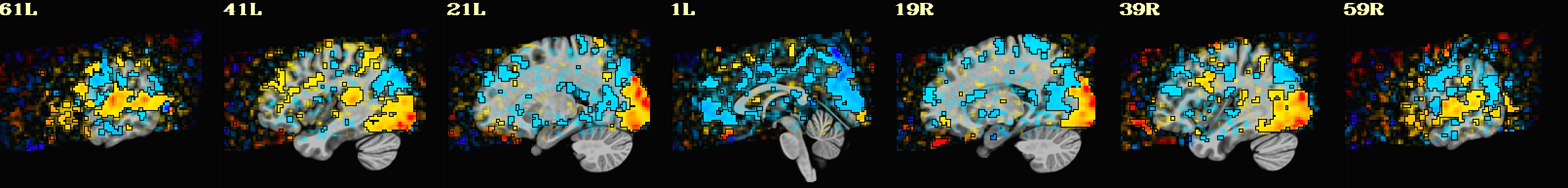
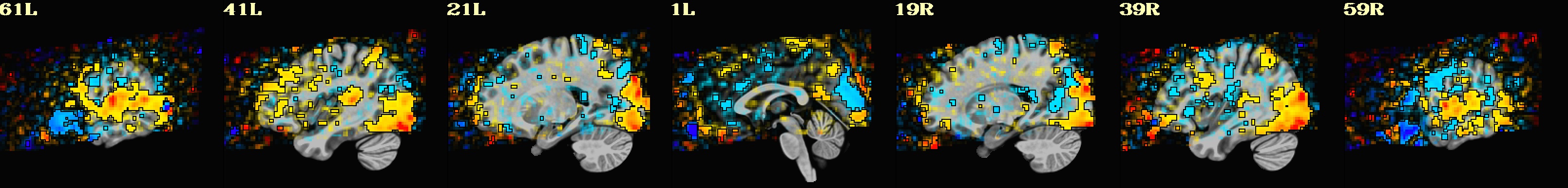
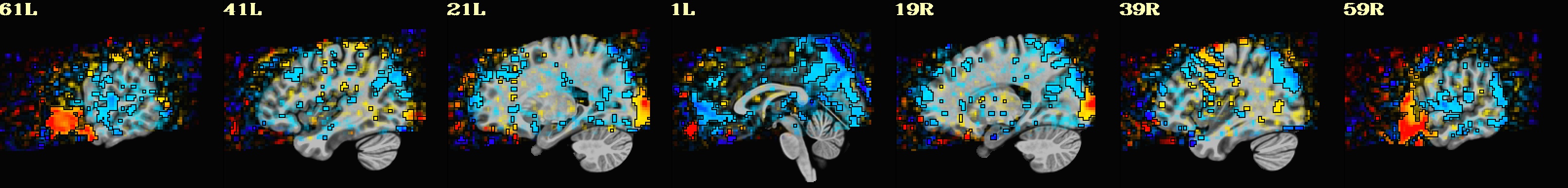
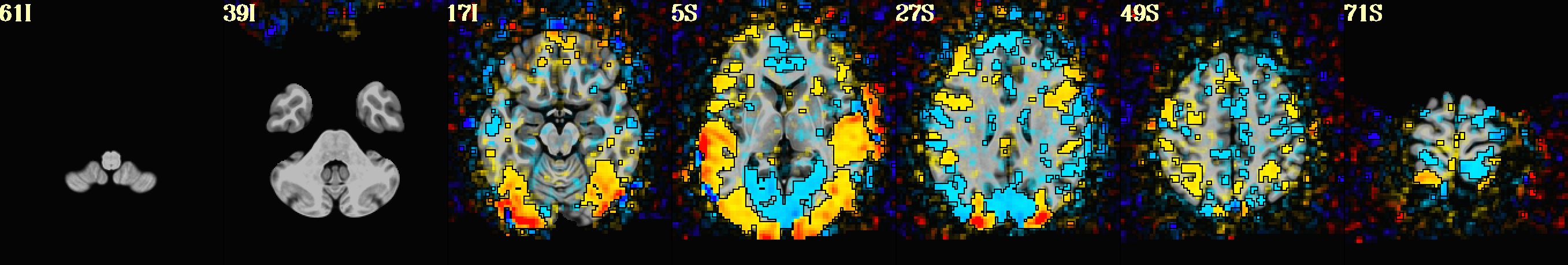
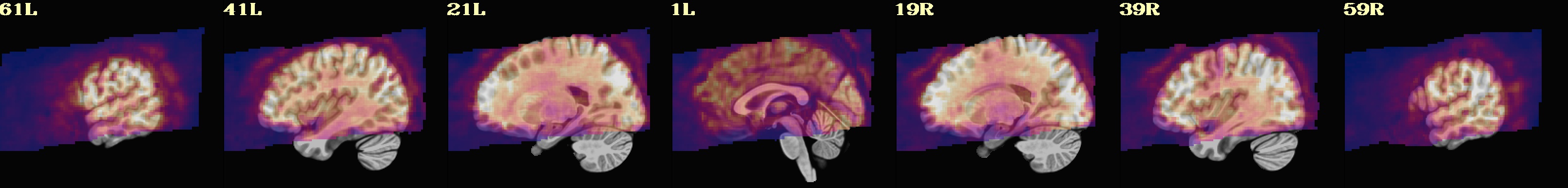
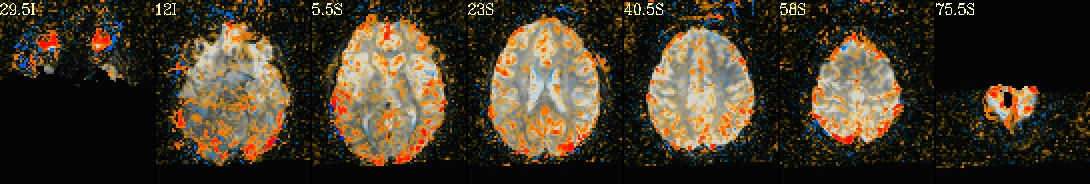
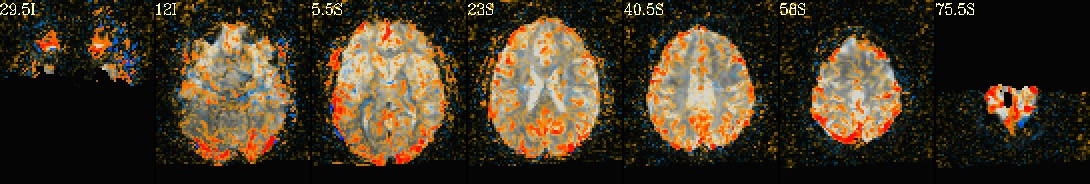
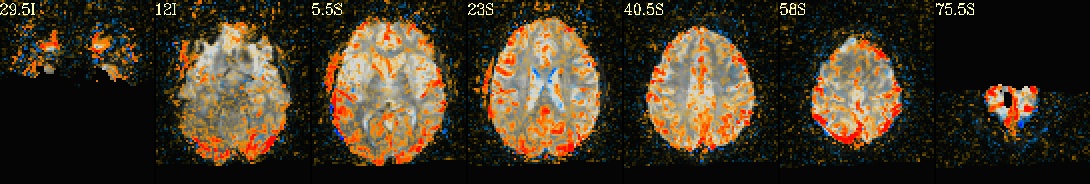
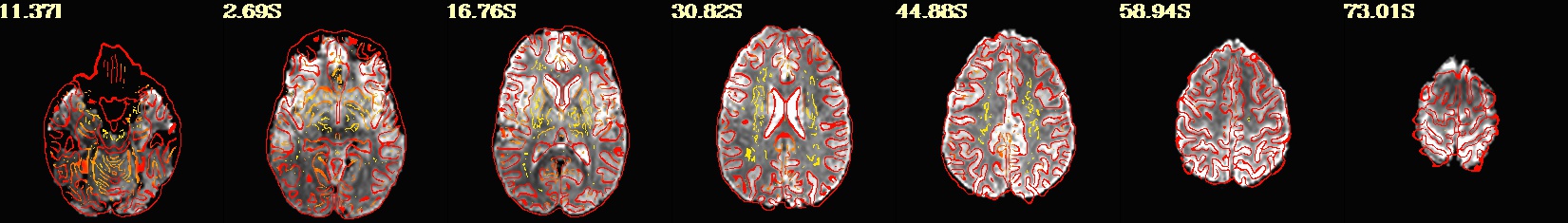
This is a demo of some features within afni_proc.py's APQC HTML. Here, you can try out the following: + QC rating+comment buttons in the top menu (click and Ctrl+click) + "NV" buttons by each dset image, to toggle NiiVue on/off and explore the datasets interactively (click and scroll; use "c" over the rendered volume to clip it.) + the "HELP" button in the top menu to see more features. "AV", "IC" and "GV" buttons work on computers with AFNI installed. See details in: https://doi.org/10.1101/2024.03.27.586976
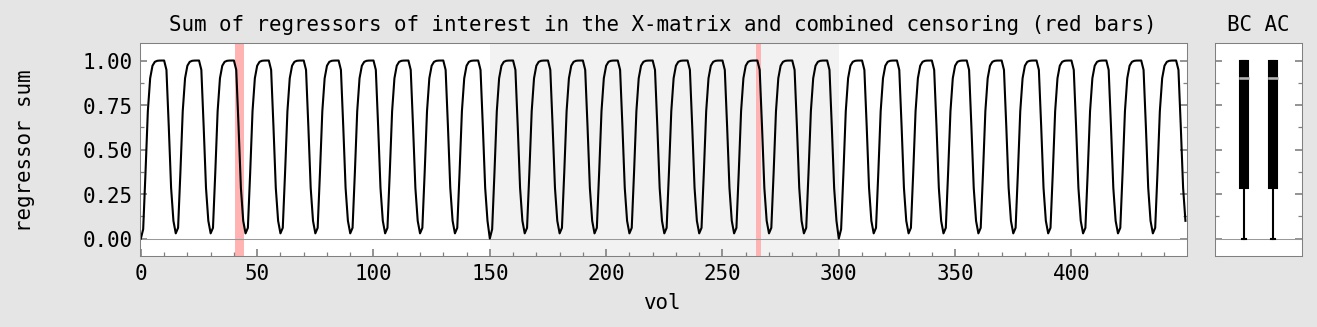
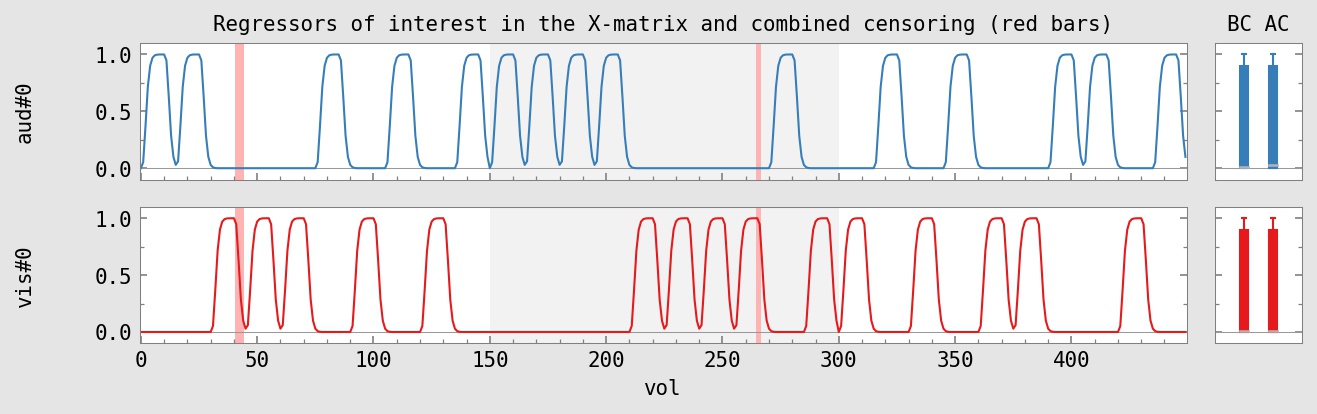
initial DF : 450 : 100.0% DF used for regs of interest : 2 : 0.4% DF used for censoring : 6 : 1.3% DF used for polort : 12 : 2.7% DF used for motion : 18 : 4.0% total DF used : 38 : 8.4% final DF : 412 : 91.6%
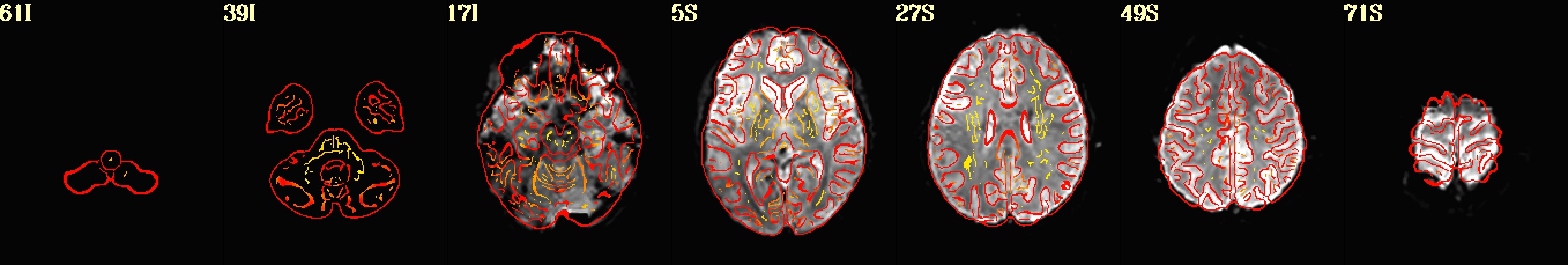
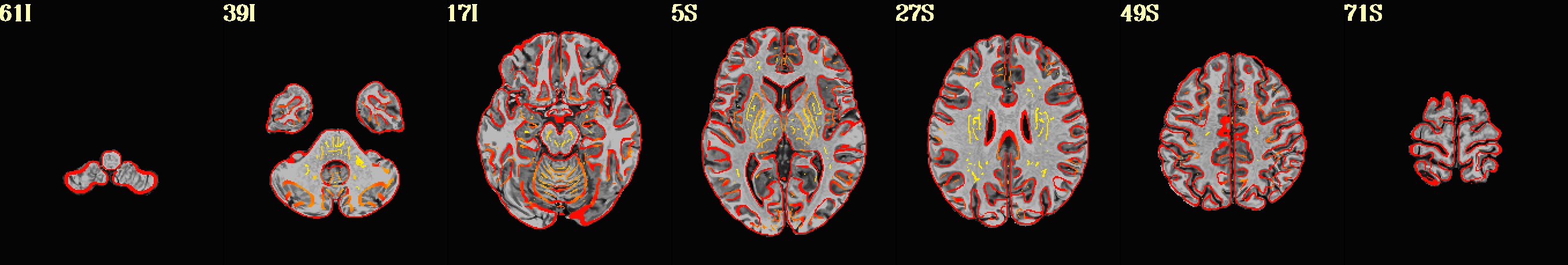
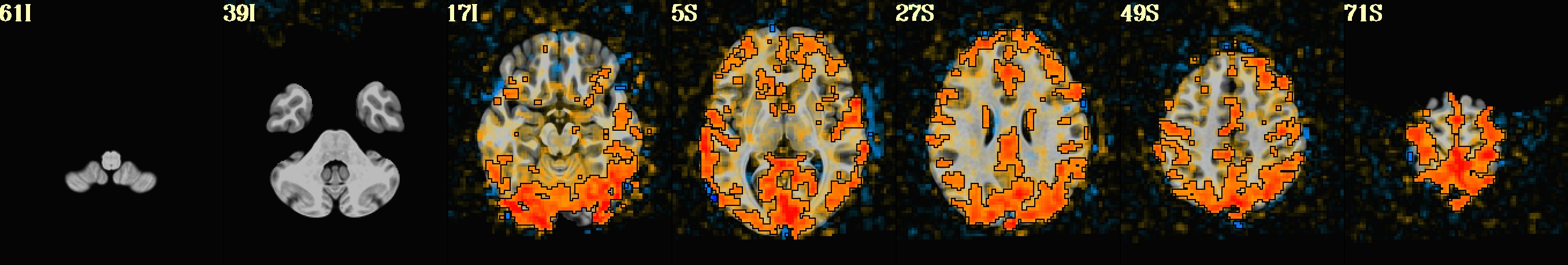
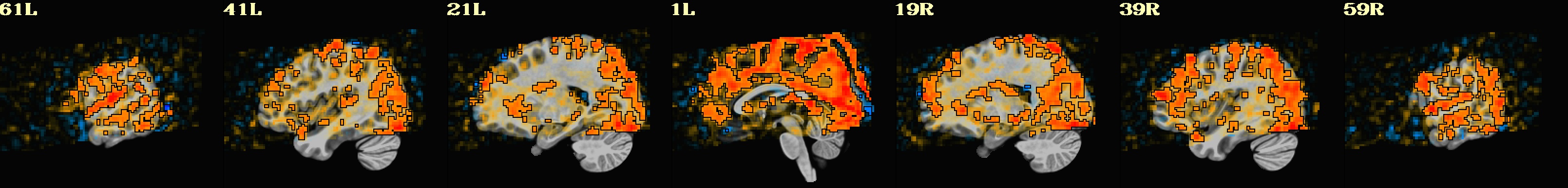
dset: MNI_2009c_asym (ROI_import_MNI_2009c_asym_resam), Nroi: 12 ---------------------------------------------------------------- ROI Nvox Nzer Dvox Tmin T25% Tmed T75% Tmax X Y Z ROI_name --- ---- ---- ---- ---- ---- ---- ---- ---- ----- ----- ----- -------- 10 448 8 2.2 44 143 206 245 328 5.0 91.8 0.5 SYAF-LH_VisCent_Striate_1 11 1071 4 2.8 29 182 237 274 452 42.5 16.8 55.5 SYAF-LH_SomMotA_1 12 794 0 2.4 65 121 159 200 305 7.5 -20.8 33.0 SYAF-LH_SalVentAttnB_PFCmp_1 13 1337 5 3.0 5 18 40 82 325 15.0 -23.2 -22.0 SYAF-LH_LimbicB_OFC_1 14 1971 1345 3.0 8 50 73 105 185 27.5 11.8 -29.5 SYAF-LH_LimbicA_TempPole_1 20 839 15 2.2 9 130 179 212 330 -10.0 66.8 10.5 GHCP-R_Primary_Visual_Cortex 21 282 15 1.4 67 171 220 265 405 -7.5 34.2 78.0 GHCP-R_Primary_Sensory_Cortex 22 92 0 1.7 108 174 203 240 314 -5.0 -25.8 28.0 GHCP-R_Anterior_24_prime 23 342 0 2.2 8 15 21 28 93 -10.0 -35.8 -24.5 GHCP-R_Orbital_Frontal_Complex 24 660 433 2.2 15 42 62 110 271 -45.0 -15.8 -34.5 GHCP-R_Area_TG_dorsal 30 180 11 2.2 10 27 47 76 140 -22.5 -0.8 -24.5 JULI-RH_Amygdala 31 34 0 1.0 62 166 195 217 237 -5.0 -8.2 -4.5 JULI-RH_Accumbens
mild
Warnings regarding Correlation Matrix: X.xmat.1D severity correlation cosine regressor pair -------- ----------- ------ ---------------------------------------- medium: -0.612 0.000 (12 vs. 13) vis#0 vs. aud#0
none
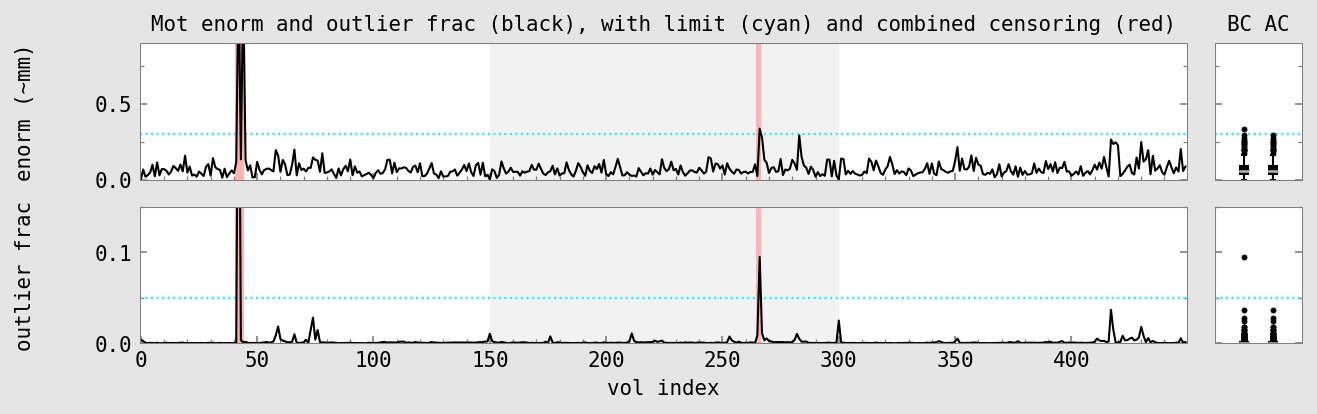
Censored 6 of 450 total time points : 1.3%
none
Max indiv stim censoring fraction : 2.5% -------------------------------------------- Censored 6 of 240 TRs of 'vis#0' stim : 2.5% Censored 0 of 239 TRs of 'aud#0' stim : 0.0%
none
none
none
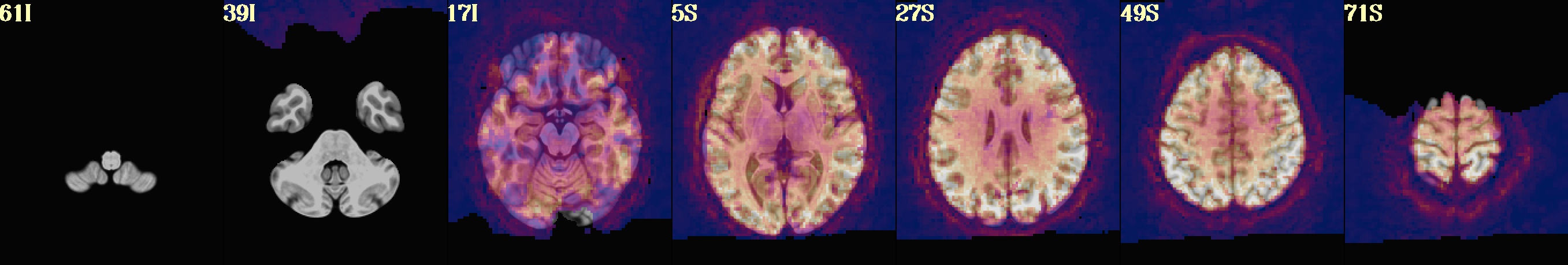
flip guess cost func original val flipped val ---------- --------- ------------ ----------- NO_FLIP lpc+ZZ -0.40431 -0.18269
medium
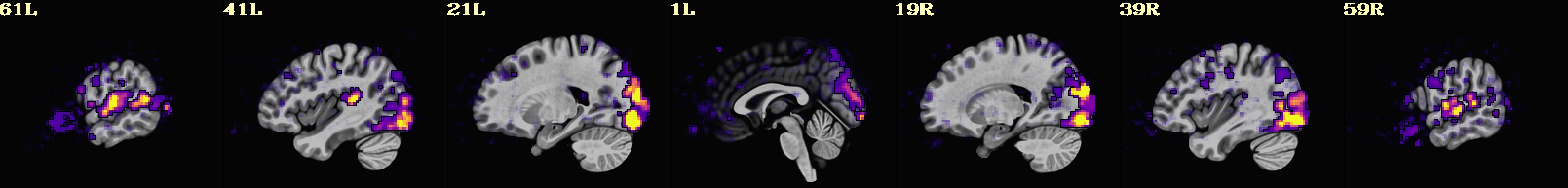
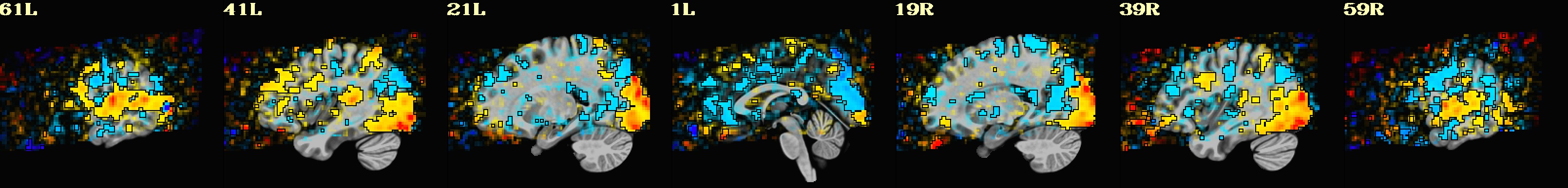
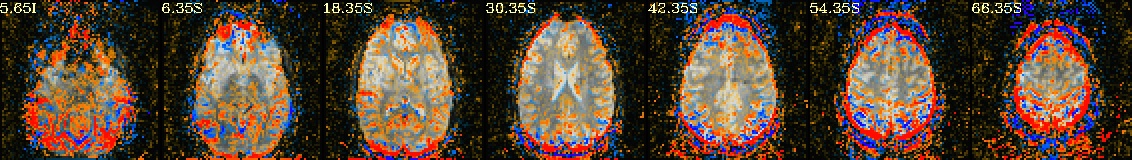
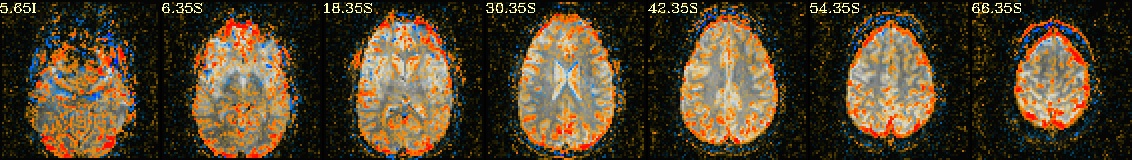
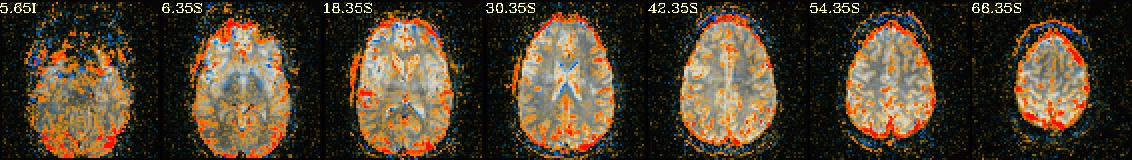
Lines per run : 4 3 5 Intersecting : 3 Coordinates (see images of the first 7, below, check locations with InstaCorr) ------------------------------------------------------------------------------ Intersecting all r01 r02 ... ----------------------- ----------------------- ----------------------- -2.70 -40.30 30.35 -2.70 -40.30 30.35 -2.70 -40.30 30.35 -41.20 -15.50 30.35 -41.20 -12.80 30.35 -41.20 -15.50 30.35 -8.20 -65.00 30.35 -8.20 -65.00 30.35 -8.20 -65.00 30.35 -27.40 -59.50 30.35
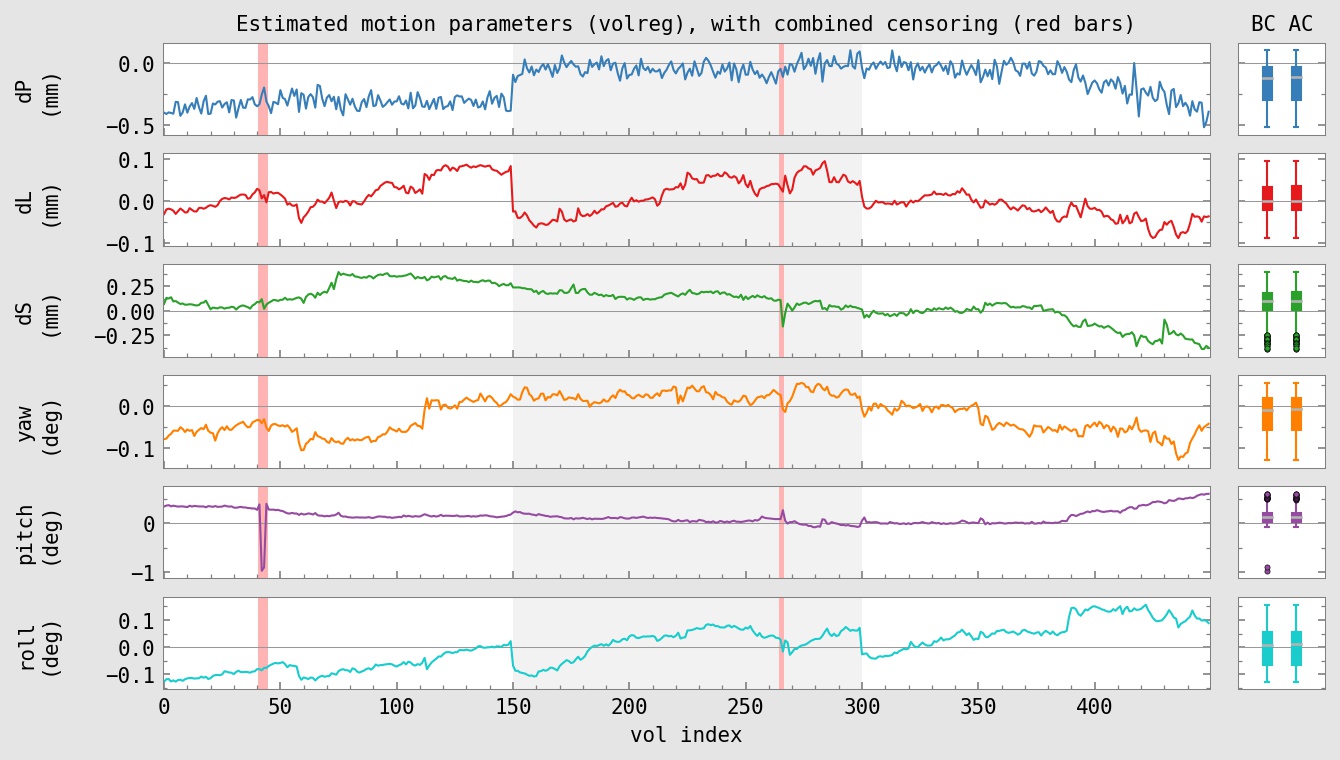
subject ID : sub-002 AFNI version : AFNI_24.0.14 AFNI package : linux_ubuntu_16_64_glw_local_shared TR : 2.0 TRs removed (per run) : 2 multiband level : 1 slice timing pattern : alt+z num stim classes provided : 2 final anatomy dset : anat_final.sub-002+tlrc.HEAD final stats dset : stats.sub-002+tlrc.HEAD orig voxel counts : 80 80 33 orig voxel resolution : 2.750000 2.750000 3.000000 orig volume center : -4.037201 -14.153702 30.349419 final voxel resolution : 2.500000 2.500000 2.500000 motion limit : 0.3 num TRs above mot limit : 3 average motion (per TR) : 0.0755233 average censored motion : 0.0691518 max motion displacement : 1.66201 max censored displacement : 1.02929 outlier limit : 0.05 average outlier frac (TR) : 0.00276734 num TRs above out limit : 2 flip guess : NO_FLIP num runs found : 3 num TRs per run : 150 150 150 num TRs per run (applied) : 146 148 150 num TRs per run (censored): 4 2 0 fraction censored per run : 0.0266667 0.0133333 0 TRs total (uncensored) : 450 TRs total : 444 degrees of freedom used : 32 degrees of freedom left : 412 final DF fraction : 0.915556 TRs censored : 6 censor fraction : 0.013333 num regs of interest : 2 num TRs per stim (orig) : 240 239 num TRs censored per stim : 6 0 fraction TRs censored : 0.025 0.000 ave mot per sresp (orig) : 0.078370 0.071256 ave mot per sresp (cens) : 0.066435 0.071256 TSNR average : 201.632 global correlation (GCOR) : 0.0487375 anat/EPI mask Dice coef : 0.852987 anat/templ mask Dice coef : 0.995383 maximum F-stat (masked) : 952.431 blur estimates (ACF) : 0.726198 3.10233 10.9184 blur estimates (FWHM) : 0 0 0